Due to the inaccessibility of database contents in several online PTM resources, a total 41 biological databases related to PTMs are integrated in dbPTM. To solve the heterogeneity among the data collected from different sources, the reported modification sites are mapped to the UniProtKB protein entries using sequence comparison. With the high-throughput of mass spectrometry-based methods in post-translational proteomics, this update also includes manually curated MS/MS-identified peptides associated with PTMs from research articles through a literature survey. First, a table list of PTM-related keywords is constructed by referring to the UniProtKB PTM list (http://www.uniprot.org/docs/ptmlist.txt) and the annotations of RESID. Then, all fields in the PubMed database are searched based on the keywords of the constructed table list. This is then followed by downloading the full text of the research articles. For the various experiments of proteomic identification, a text-mining system is developed to survey full-text literature that potentially describes the site-specific identification of modified sites.Furthermore, in order to determine the locations of PTMs on a full-length protein sequence, the experimentally verified MS/MS peptides are then mapped to UniProtKB protein entries based on its database identifier (ID) and sequence identity. In the process of data mapping, MS/MS peptides that cannot align exactly to a protein sequence are discarded. Finally, each mapped PTM site is attributed with a corresponding literature (PubMed ID). All types of PTM were categorized by the modified amino acid, with tab-delimited format. These datasets provide UniProt ID, modified position, PTM type, and the sequence with upstream 10 amino acids to downstream 10 amino acids. However, some types of PTM, which were occurred in N-terminal or C-terminal protein, were extracted the sequences with dashes ('-').
| PTM Type | Number of experimental Sites | Number of literatures | Download | |
|---|---|---|---|---|
| ADP-ribosylation | 107 | 35 | Windows | MAC / Linux |
| AMPylation | 27 | 20 | Windows | MAC / Linux |
| Acetylation | 138169 | 19451 | Windows | MAC / Linux |
| Amidation | 3316 | 1084 | Windows | MAC / Linux |
| Biotinylation | 12 | 12 | Windows | MAC / Linux |
| Blocked amino end | 26 | 25 | Windows | MAC / Linux |
| Butyrylation | 82 | 3 | Windows | MAC / Linux |
| C-linked Glycosylation | 211 | 19 | Windows | MAC / Linux |
| Carbamidation | 22 | 1 | Windows | MAC / Linux |
| Carboxyethylation | 1 | 1 | Windows | MAC / Linux |
| Carboxylation | 44 | 63 | Windows | MAC / Linux |
| Cholesterol ester | 2 | 2 | Windows | MAC / Linux |
| Citrullination | 122 | 18 | Windows | MAC / Linux |
| Crotonylation | 373 | 7 | Windows | MAC / Linux |
| D-glucuronoylation | 1 | 1 | Windows | MAC / Linux |
| Deamidation | 92 | 44 | Windows | MAC / Linux |
| Deamination | 42 | 13 | Windows | MAC / Linux |
| Decanoylation | 6 | 7 | Windows | MAC / Linux |
| Decarboxylation | 2 | 2 | Windows | MAC / Linux |
| Dephosphorylation | 133 | 95 | Windows | MAC / Linux |
| Disulfide bond | 6 | 1 | Windows | MAC / Linux |
| Farnesylation | 87 | 83 | Windows | MAC / Linux |
| Formation of an isopeptide bond | 29 | 16 | Windows | MAC / Linux |
| Formylation | 256 | 47 | Windows | MAC / Linux |
| GPI-anchor | 103 | 57 | Windows | MAC / Linux |
| Gamma-carboxyglutamic acid | 508 | 94 | Windows | MAC / Linux |
| Geranylgeranylation | 81 | 48 | Windows | MAC / Linux |
| Glutarylation | 952 | 4 | Windows | MAC / Linux |
| Glutathionylation | 4129 | 85 | Windows | MAC / Linux |
| Hydroxyceramide ester | 3 | 1 | Windows | MAC / Linux |
| Hydroxylation | 2404 | 353 | Windows | MAC / Linux |
| Iodination | 19 | 2 | Windows | MAC / Linux |
| Lactoylation | 306 | 1 | Windows | MAC / Linux |
| Lactylation | 336 | 2 | Windows | MAC / Linux |
| Lipoylation | 35 | 30 | Windows | MAC / Linux |
| Malonylation | 12847 | 12 | Windows | MAC / Linux |
| Methylation | 16114 | 8897 | Windows | MAC / Linux |
| Myristoylation | 312 | 207 | Windows | MAC / Linux |
| N-carbamoylation | 1 | 1 | Windows | MAC / Linux |
| N-linked Glycosylation | 27361 | 1941 | Windows | MAC / Linux |
| N-palmitoylation | 74 | 64 | Windows | MAC / Linux |
| Neddylation | 1676 | 4 | Windows | MAC / Linux |
| Nitration | 81 | 17 | Windows | MAC / Linux |
| O-linked Glycosylation | 16692 | 4457 | Windows | MAC / Linux |
| O-palmitoleoylation | 6 | 11 | Windows | MAC / Linux |
| O-palmitoylation | 3 | 3 | Windows | MAC / Linux |
| Octanoylation | 7 | 8 | Windows | MAC / Linux |
| Oxidation | 427 | 83 | Windows | MAC / Linux |
| Phosphatidylethanolamine amidation | 9 | 7 | Windows | MAC / Linux |
| Phosphorylation | 1615054 | 44986 | Windows | MAC / Linux |
| Propionylation | 13 | 3 | Windows | MAC / Linux |
| Pyrrolidone carboxylic acid | 965 | 571 | Windows | MAC / Linux |
| Pyrrolylation | 1 | 1 | Windows | MAC / Linux |
| Pyruvate | 25 | 21 | Windows | MAC / Linux |
| S-Cyanation | 1 | 2 | Windows | MAC / Linux |
| S-archaeol | 1 | 1 | Windows | MAC / Linux |
| S-carbamoylation | 1 | 2 | Windows | MAC / Linux |
| S-cysteinylation | 2 | 2 | Windows | MAC / Linux |
| S-diacylglycerol | 58 | 50 | Windows | MAC / Linux |
| S-linked Glycosylation | 5 | 7 | Windows | MAC / Linux |
| S-nitrosylation | 4172 | 190 | Windows | MAC / Linux |
| S-palmitoylation | 6501 | 571 | Windows | MAC / Linux |
| Serotonylation | 9 | 2 | Windows | MAC / Linux |
| Stearoylation | 3 | 3 | Windows | MAC / Linux |
| Succinylation | 17973 | 59 | Windows | MAC / Linux |
| Sulfation | 252 | 129 | Windows | MAC / Linux |
| Sulfhydration | 8 | 7 | Windows | MAC / Linux |
| Sulfoxidation | 7581 | 124 | Windows | MAC / Linux |
| Sumoylation | 5889 | 221 | Windows | MAC / Linux |
| Thiocarboxylation | 13 | 13 | Windows | MAC / Linux |
| UMPylation | 10 | 8 | Windows | MAC / Linux |
| Ubiquitination | 348307 | 669 | Windows | MAC / Linux |
Owing to the labor-intensive MS/MS-based experiments, a variety of computational methods have been proposed to identify putative PTM sites based on protein sequence. With numerous PTM prediction methods, it is difficult to determine a best prediction tool merely according to their cross-validation performances. Although most of these studies have provided independent testing results for their prediction methods, there is no standard dataset for the evaluation of predictive powers among various PTM prediction tools. Therefore, this update compiles non-homologous benchmark datasets to evaluate the predictive power for PTM sites prediction tools, that provides suggestions to users with the need to predict PTM sites with high sensitivity (Sn), high specificity (Sp), or balanced Sn and Sp.
| PTM Type | Number of proteins | Number of positive sites | Number of negative sites | Download | |
|---|---|---|---|---|---|
| Phosphorylation by CDK | 1,020 | 1,503 | 29,823 | Windows | MAC / Linux |
| Phosphorylation by MAPK | 857 | 1,270 | 22,436 | Windows | MAC / Linux |
| Phosphorylation by PKA | 905 | 1,209 | 29,813 | Windows | MAC / Linux |
| Phosphorylation by PKC | 691 | 943 | 24,207 | Windows | MAC / Linux |
| Phosphorylation by CK2 | 511 | 819 | 15,387 | Windows | MAC / Linux |
| Phosphorylation by CAMKL | 454 | 556 | 20,129 | Windows | MAC / Linux |
| Phosphorylation by GSK | 291 | 397 | 10,328 | Windows | MAC / Linux |
| Phosphorylation by AKT | 351 | 380 | 14,617 | Windows | MAC / Linux |
| Phosphorylation by CAMK2 | 254 | 366 | 12,575 | Windows | MAC / Linux |
| Phosphorylation by CK1 | 174 | 339 | 5,808 | Windows | MAC / Linux |
| Phosphorylation by RSK | 221 | 215 | 6,985 | Windows | MAC / Linux |
| Phosphorylation by GRK | 77 | 147 | 2,310 | Windows | MAC / Linux |
| Phosphorylation by PKG | 126 | 145 | 7,311 | Windows | MAC / Linux |
| Phosphorylation by DYRK | 109 | 142 | 3,470 | Windows | MAC / Linux |
| Phosphorylation by MAPKAPK | 100 | 125 | 3,096 | Windows | MAC / Linux |
| Phosphorylation by DMPK | 99 | 109 | 3,533 | Windows | MAC / Linux |
| Phosphorylation by PKD | 88 | 97 | 3,401 | Windows | MAC / Linux |
| Phosphorylation by PDK1 | 77 | 93 | 2,274 | Windows | MAC / Linux |
| Phosphorylation by SGK | 63 | 77 | 3,057 | Windows | MAC / Linux |
| Phosphorylation by RAD53 | 29 | 75 | 1,560 | Windows | MAC / Linux |
| Phosphorylation by DAPK | 51 | 53 | 1,284 | Windows | MAC / Linux |
| Phosphorylation by PKN | 26 | 50 | 866 | Windows | MAC / Linux |
| Phosphorylation by CAMK1 | 34 | 44 | 2,342 | Windows | MAC / Linux |
| Phosphorylation by MLCK | 20 | 34 | 484 | Windows | MAC / Linux |
| Phosphorylation by NDR | 28 | 32 | 1,096 | Windows | MAC / Linux |
| Acetylation | 5,646 | 14,407 | 8,704 | Windows | MAC / Linux |
| Citrullination | 66 | 76 | 1,501 | Windows | MAC / Linux |
| C-linked Glycosylation | 39 | 113 | 159 | Windows | MAC / Linux |
| Crotonylation | 20 | 117 | 36 | Windows | MAC / Linux |
| Formylation | 130 | 172 | 1,452 | Windows | MAC / Linux |
| Gamma-carboxyglutamic acid | 54 | 319 | 553 | Windows | MAC / Linux |
| Glutarylation | 217 | 725 | 2,543 | Windows | MAC / Linux |
| Glutathionylation | 1,493 | 3,555 | 6,617 | Windows | MAC / Linux |
| Hydroxylation | 201 | 1,270 | 2,900 | Windows | MAC / Linux |
| Lipoylation | 28 | 29 | 779 | Windows | MAC / Linux |
| Malonylation | 2,768 | 7,635 | 17,371 | Windows | MAC / Linux |
| Methylation | 5,438 | 14,686 | 36,501 | Windows | MAC / Linux |
| Nitration | 61 | 64 | 983 | Windows | MAC / Linux |
| N-linked Glycosylation | 1,969 | 2,517 | 8,330 | Windows | MAC / Linux |
| O-linked Glycosylation | 1,298 | 4,470 | 37,969 | Windows | MAC / Linux |
| S-diacylglycerol | 23 | 57 | 59 | Windows | MAC / Linux |
| S-nitrosylation | 1,434 | 3,592 | 5,803 | Windows | MAC / Linux |
| Succinylation | 2,599 | 5,049 | 5,526 | Windows | MAC / Linux |
| Sumoylation | 1,432 | 5,191 | 16,066 | Windows | MAC / Linux |
| Ubiquitination | 4,453 | 9,767 | 8,579 | Windows | MAC / Linux |
To advance cancer research, we present a collection of proteomics datasets spanning multiple cancer types. These datasets offer valuable insights into tumor and normal tissue samples, supporting research on the molecular mechanisms driving cancer progression. The table below lists critical details for each dataset, including the cancer type, number of tumor and normal samples, access to volcano plots for differential expression analysis, and direct download links. This resource empowers researchers to explore protein-level changes, aiding in discovering potential biomarkers and therapeutic targets.
| Cancer Type | Number of Tumor Sample | Number of Normal Sample | Volcano Plot | Download | Source |
|---|---|---|---|---|---|
| Colon Adenocarcinoma (COAD) | 97 | 100 | 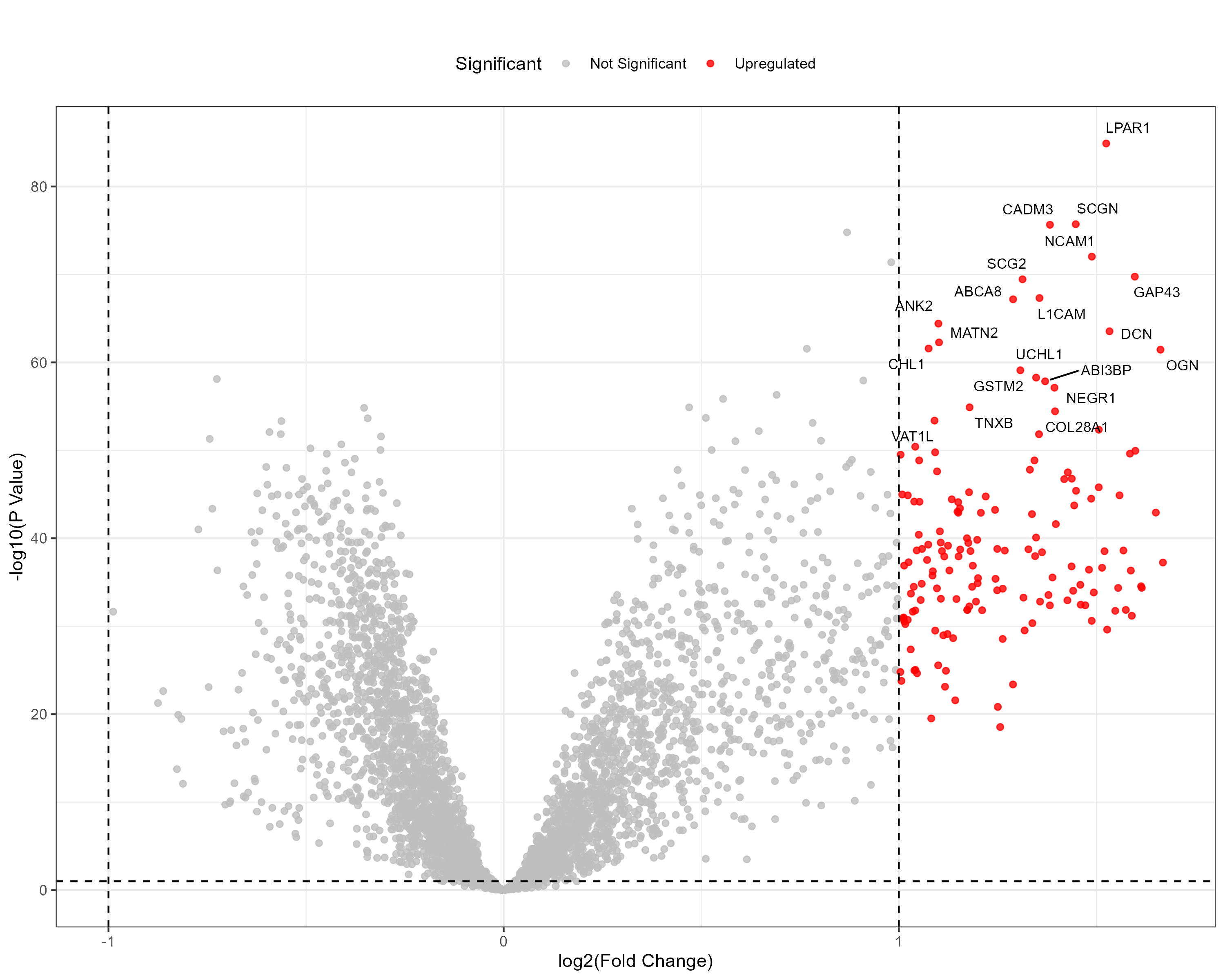
|
Windows / MAC / Linux | PDC000109 |
| Ovarian Serous Cystadenocarcinoma (OV) | 83 | 20 | 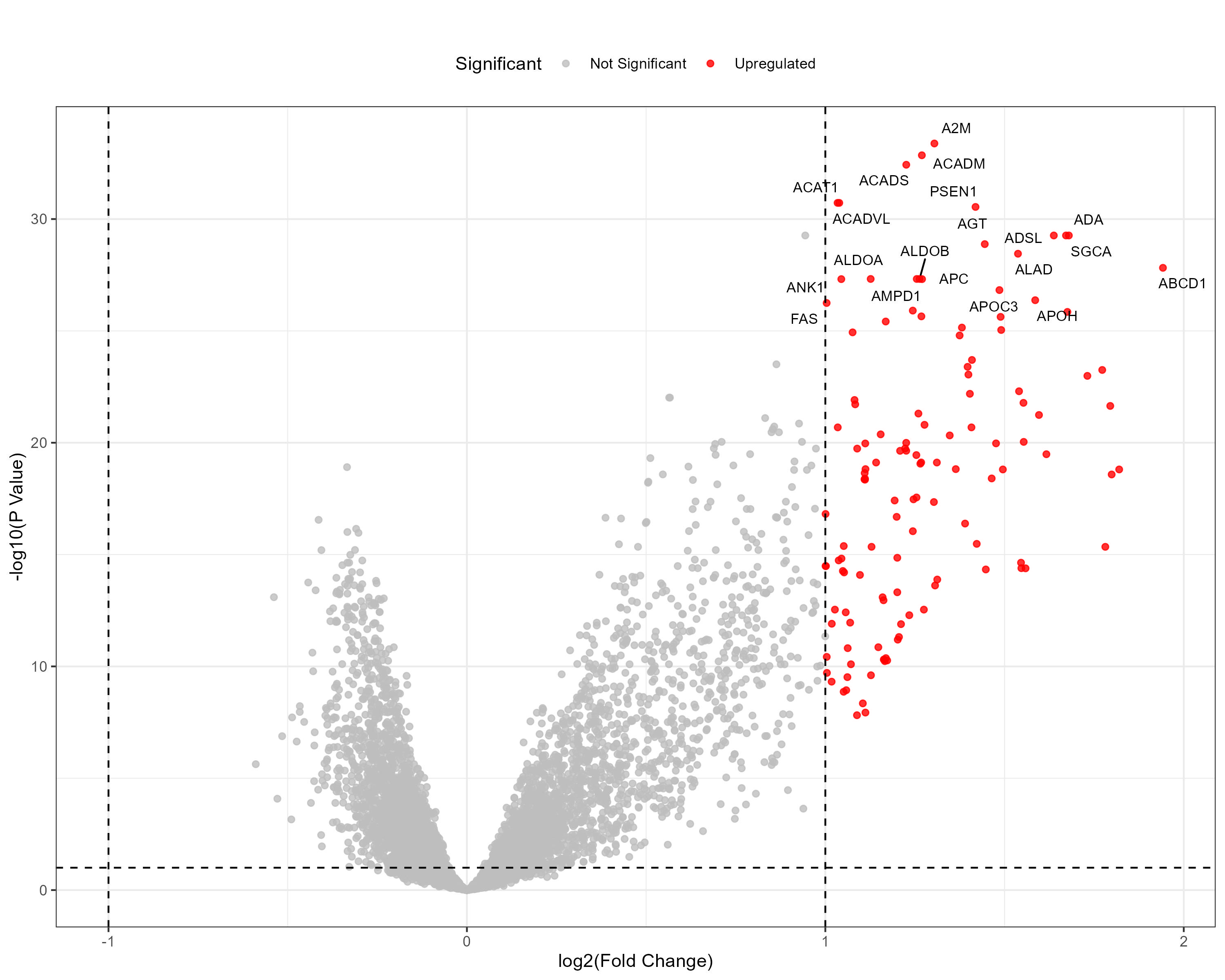
|
Windows / MAC / Linux | PDC000110 |
| Breast Invasive Carcinoma (BRCA) | 135 | 18 | 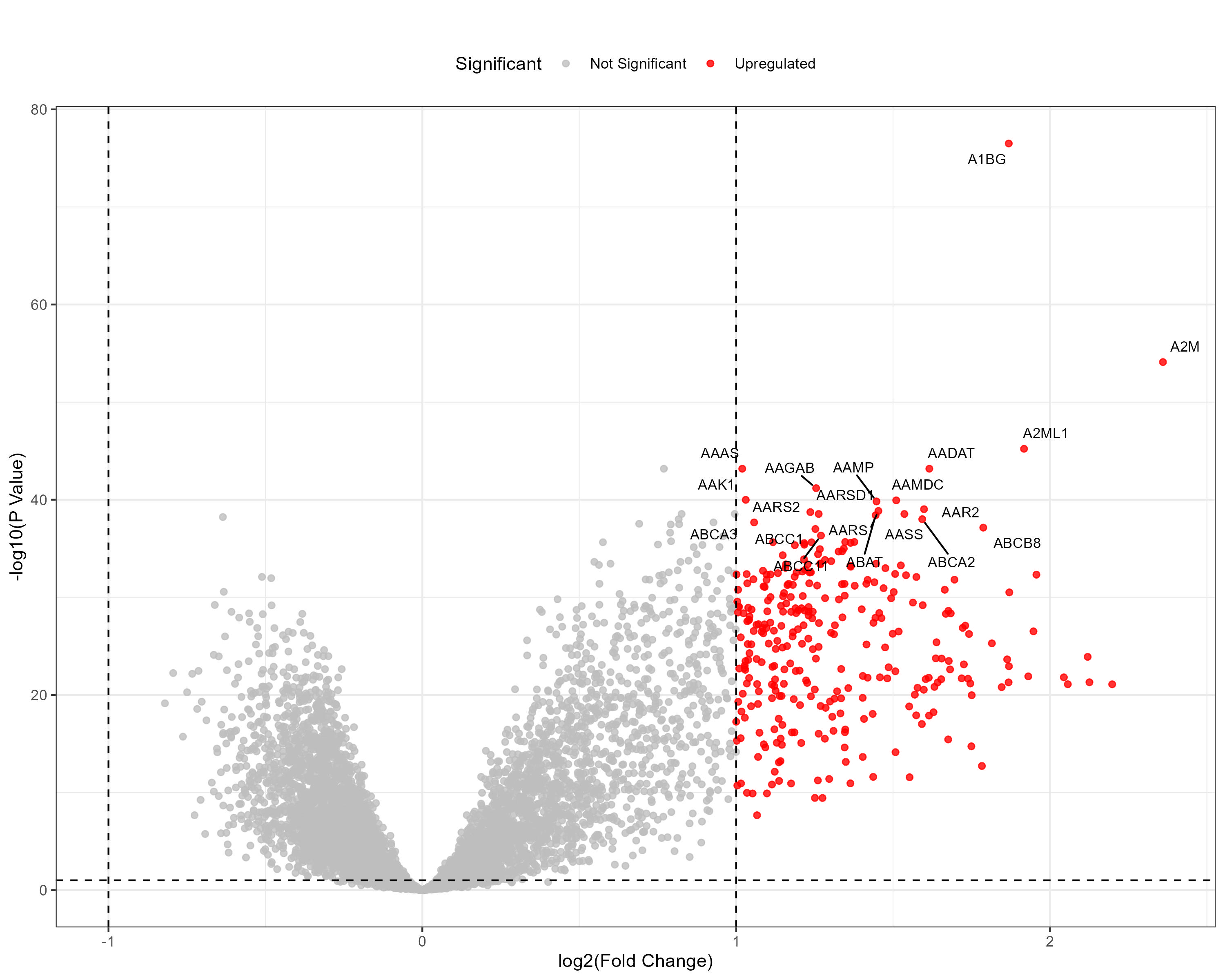
|
Windows / MAC / Linux | PDC000120 |
| Uterine Corpus Endometrial Carcinoma (UCEC) | 104 | 49 | 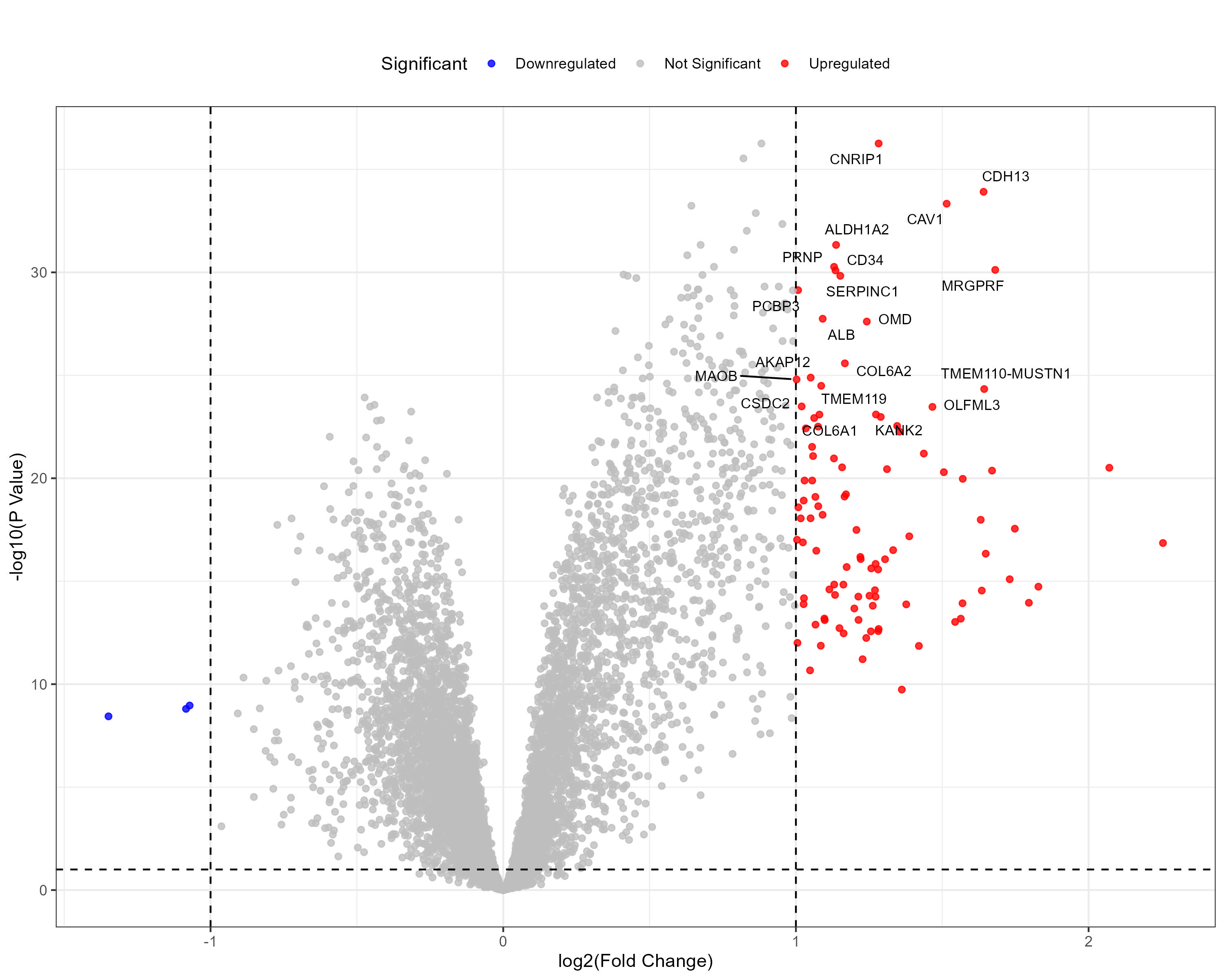
|
Windows / MAC / Linux | PDC000125 |
| Clear Cell Renal Cell Carcinoma (CCRCC) | 110 | 84 | 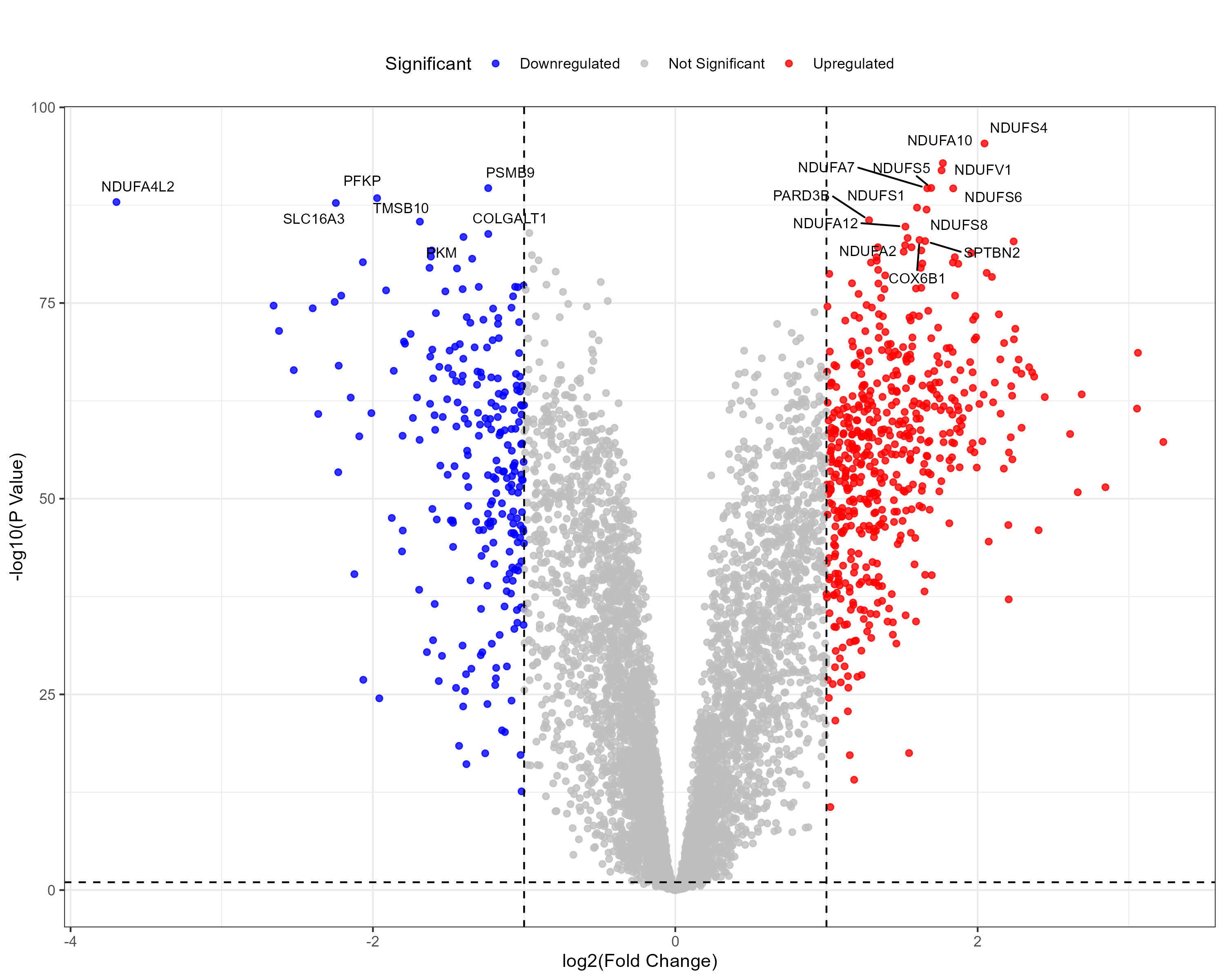
|
Windows / MAC / Linux | PDC000127 |
| Lung Adenocarcinoma (LUAD) | 110 | 101 | 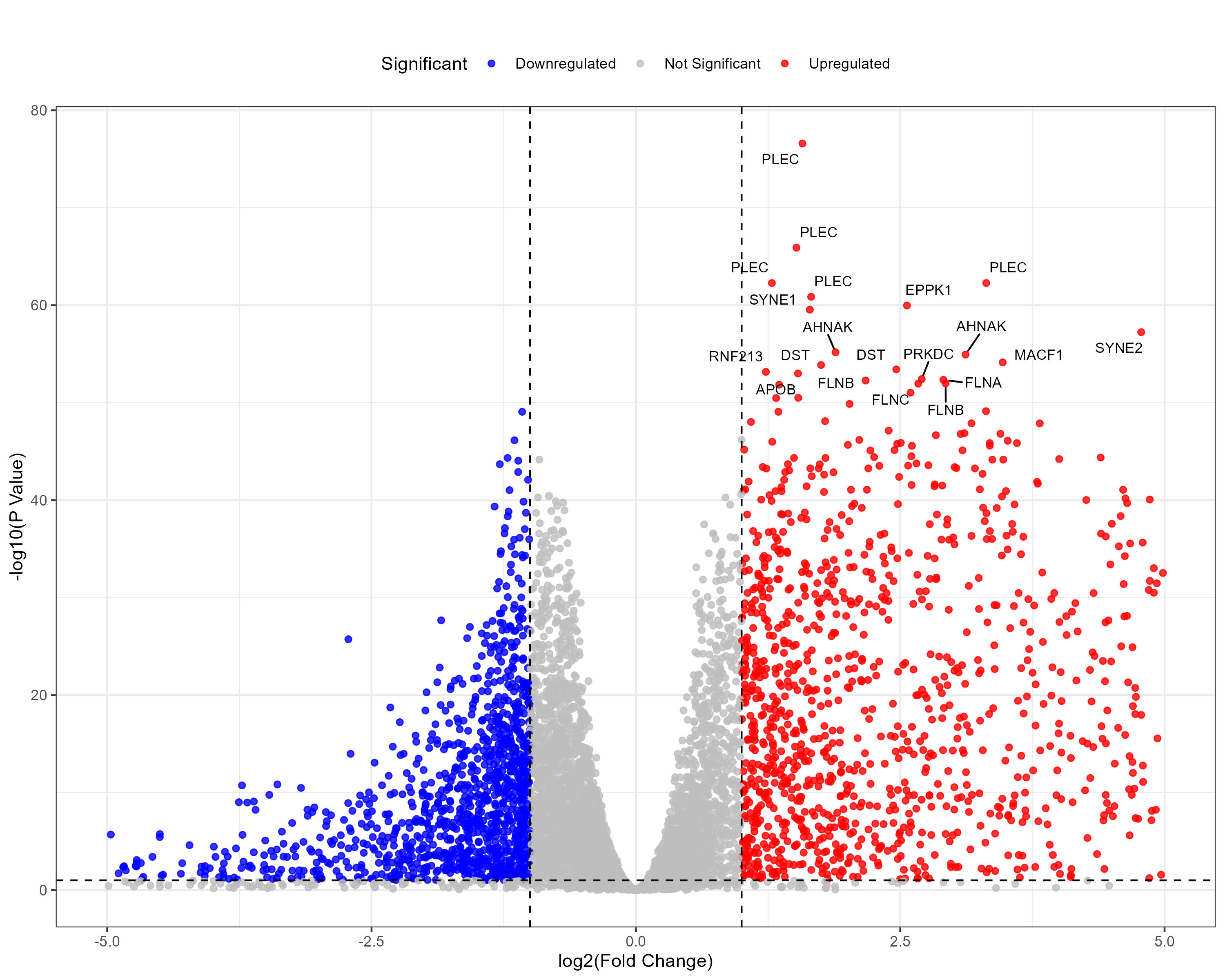
|
Windows / MAC / Linux | PDC000153 |
| Hepatocellular carcinoma (HCC) | 165 | 165 | 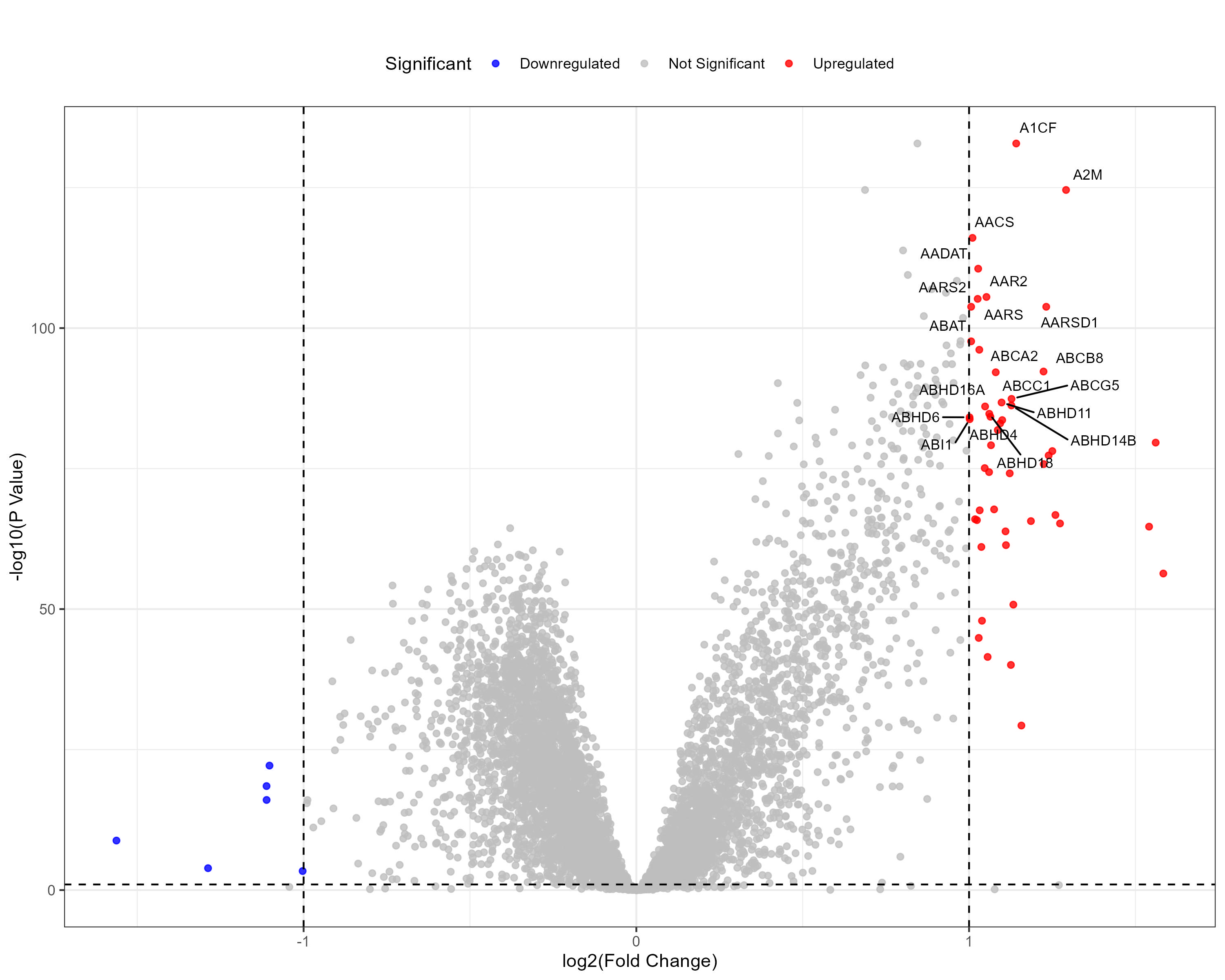
|
Windows / MAC / Linux | PDC000198 |
| Glioblastoma (GBM) | 99 | 10 | 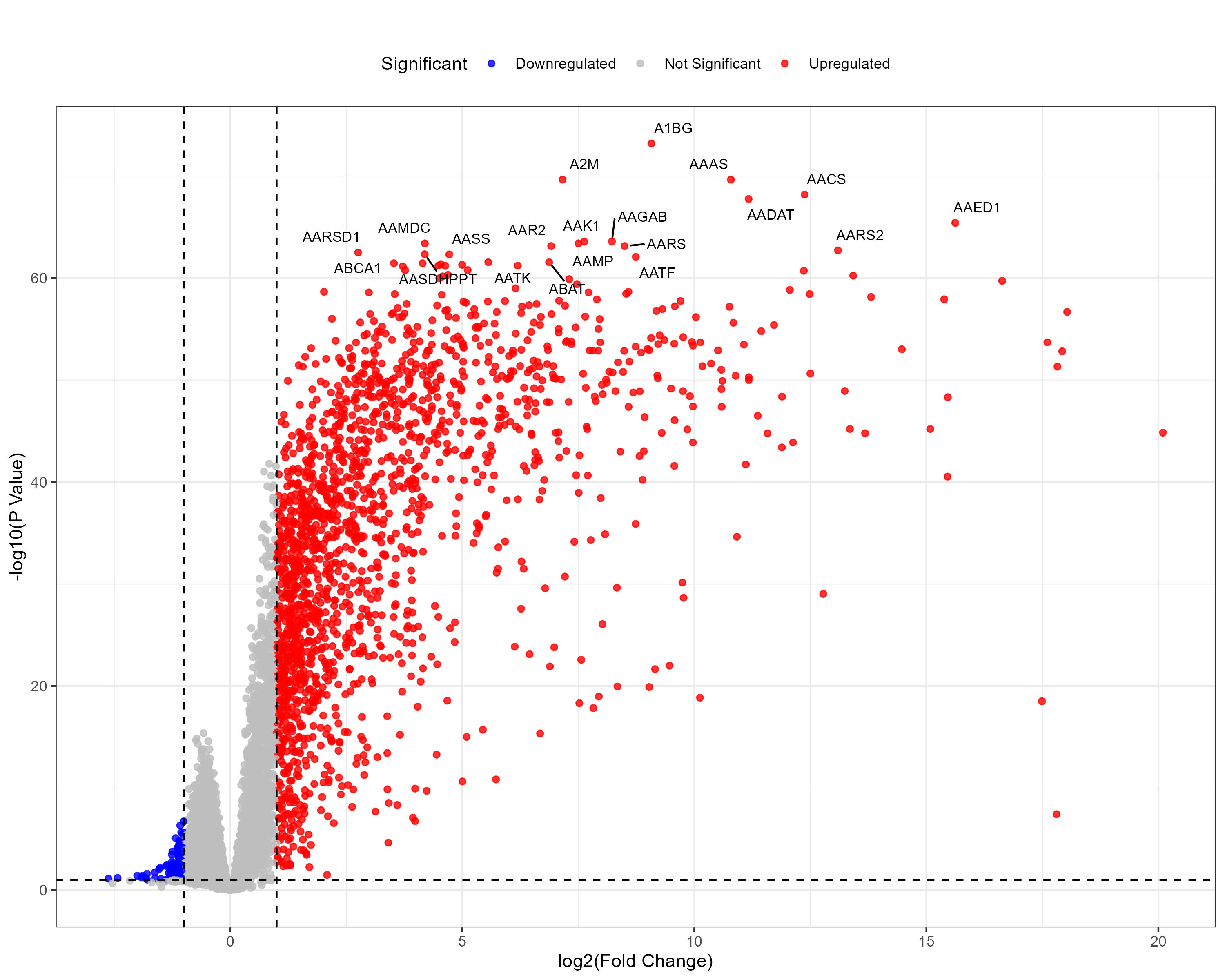
|
Windows / MAC / Linux | PDC000204 |
| Early Onset Gastric Cancer (EOGC) | 80 | 80 | 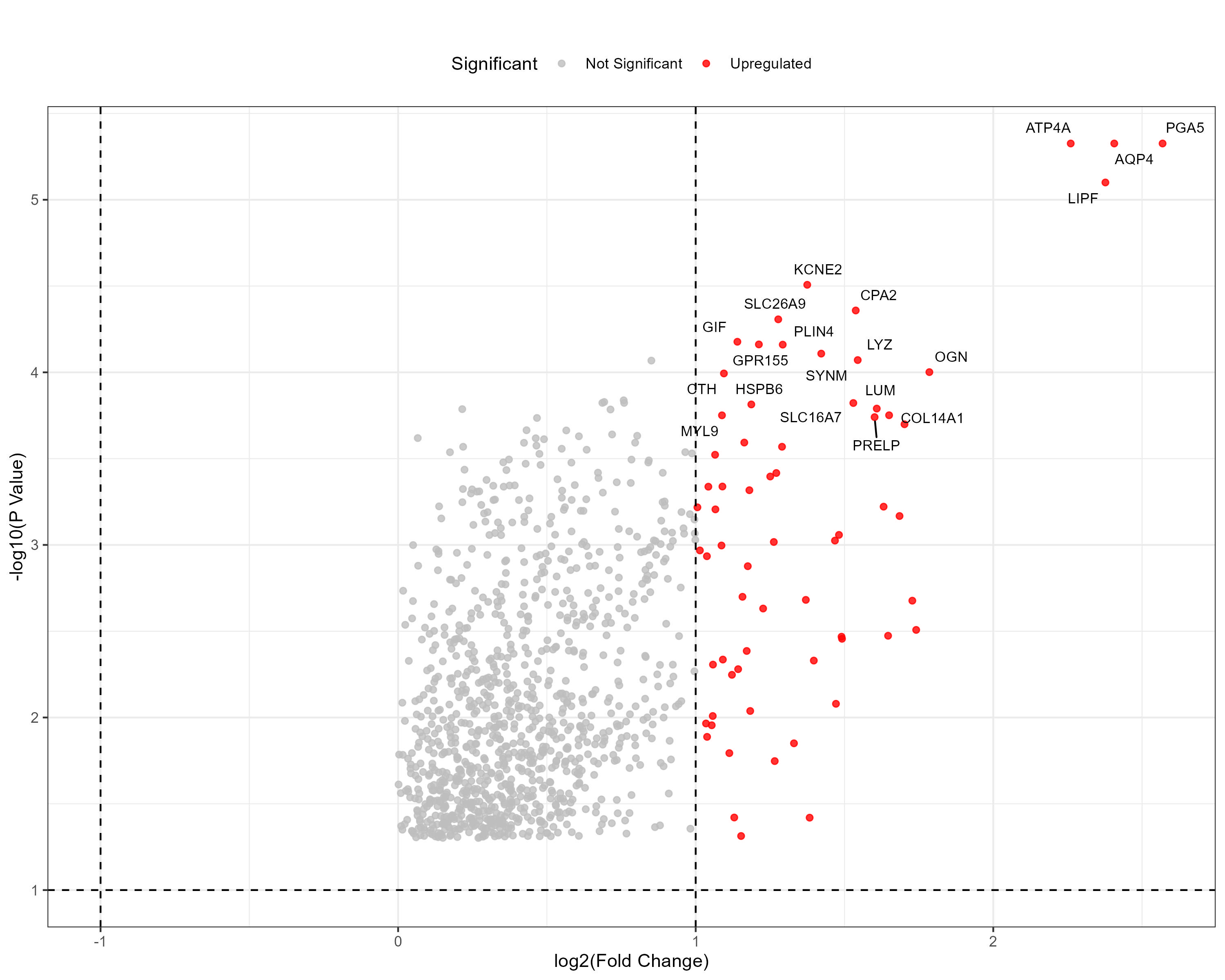
|
Windows / MAC / Linux | PDC000214 |
| Head and Neck Squamous Cell Carcinoma (HNSCC) | 109 | 63 | 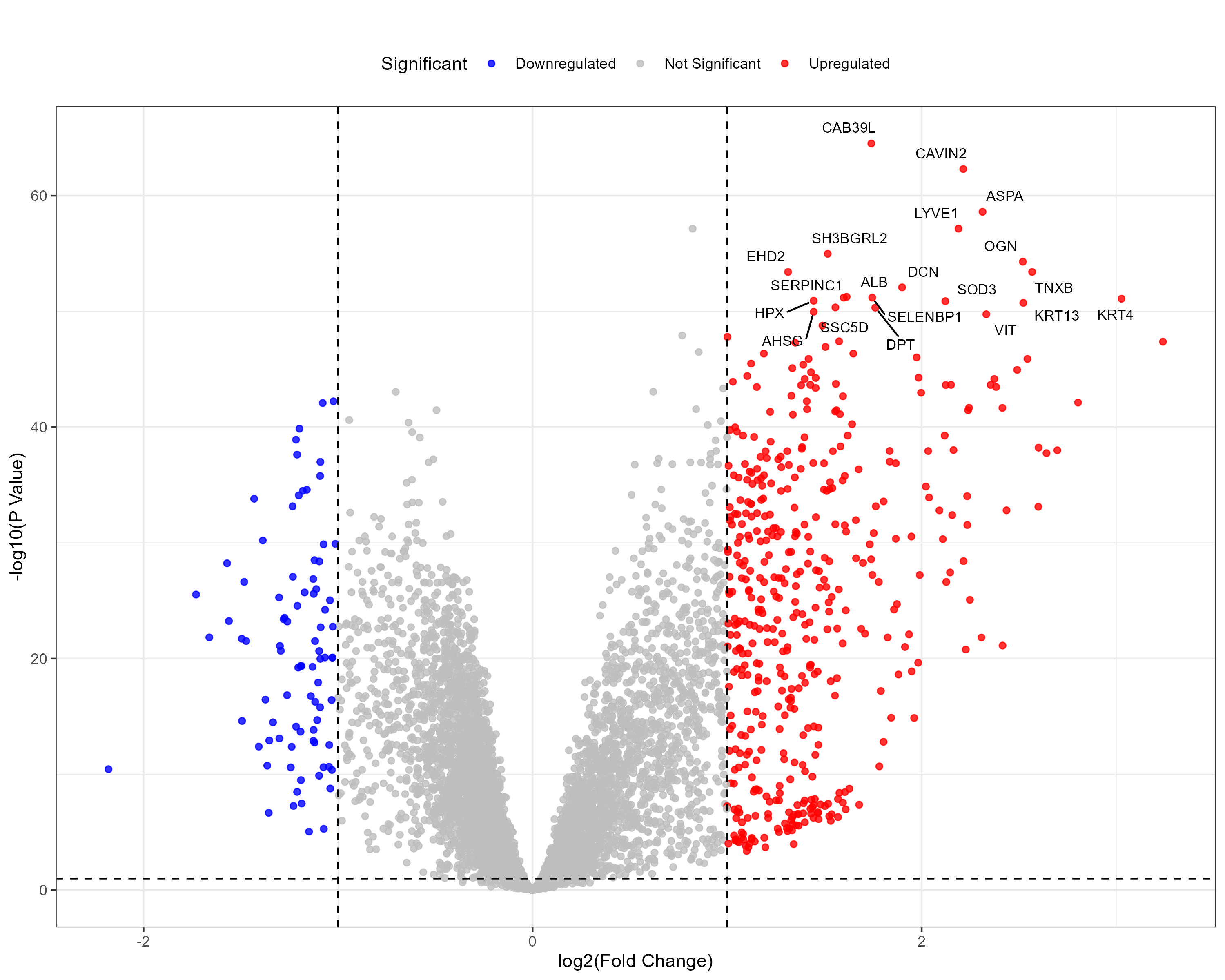
|
Windows / MAC / Linux | PDC000221 |
| Lung Squamous Cell Carcinoma (LSCC) | 108 | 99 | 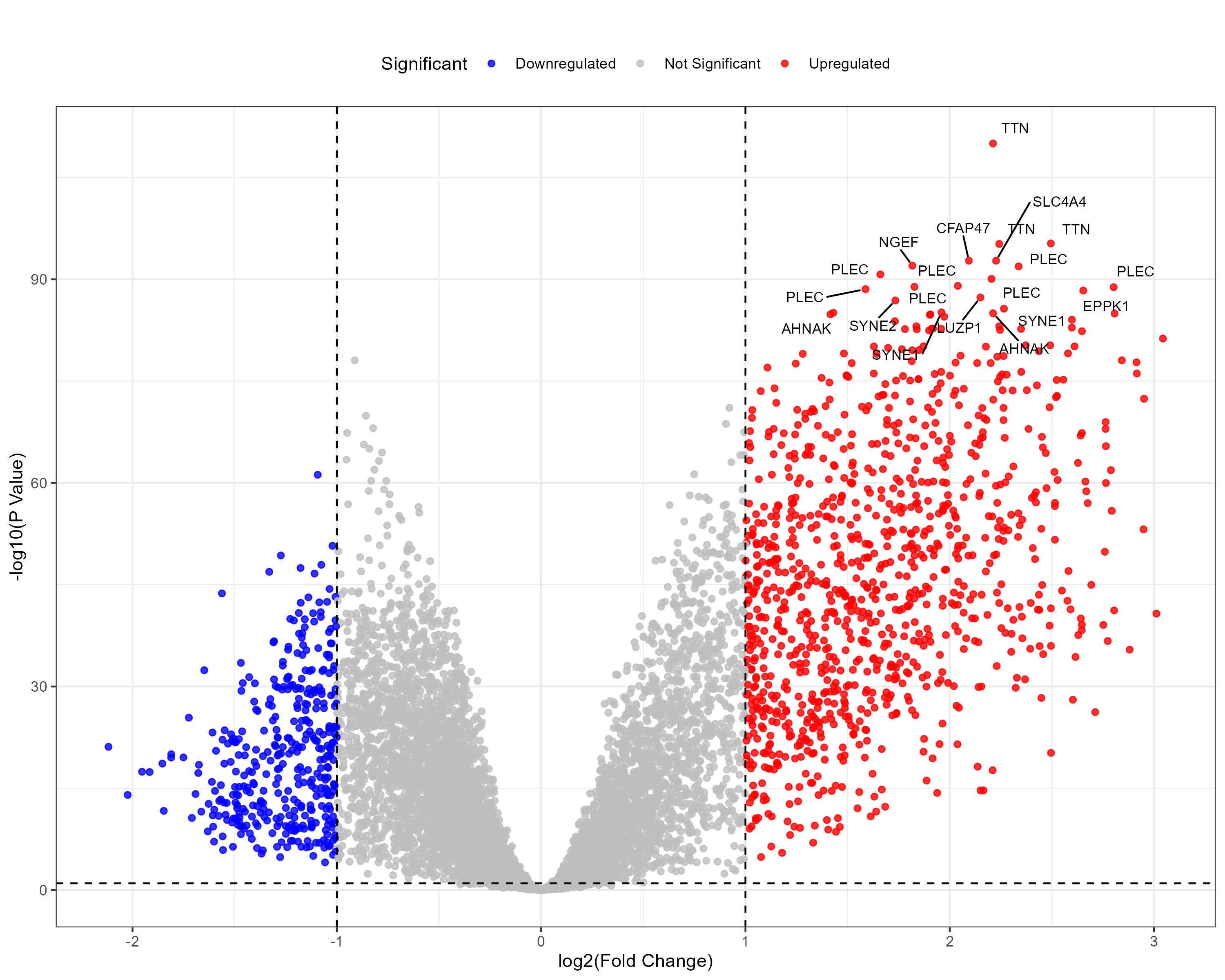
|
Windows / MAC / Linux | PDC000234 |
| Pancreatic Ductal Adenocarcinoma (PDAC) | 140 | 75 | 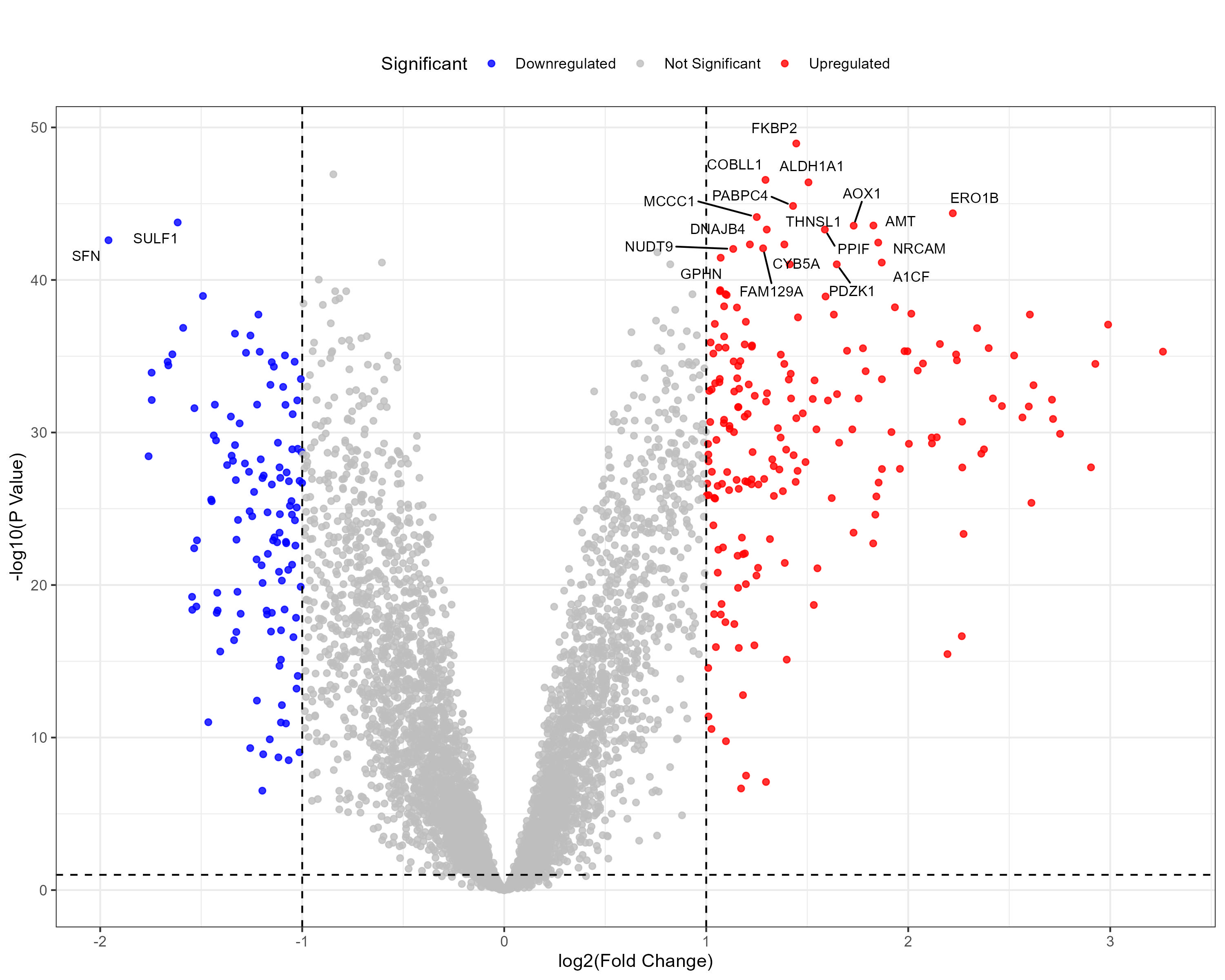
|
Windows / MAC / Linux | PDC000270 |